St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
Jiyang Yu Lab
Dissecting molecular (re)wiring and underpinning hidden drivers within and between cells/systems toward clinical translation
About the lab
Biological processes operate through molecular networks at the cellular level and cell–cell communication networks at the tissue/organ level. Deciphering the wiring and rewiring of these networks under normal and pathologic conditions is a fundamental goal of biomedical research. Our lab is focused on developing data-driven systems biology algorithms to integrate bulk, single-cell, and spatial omics data to decipher these (re)wiring events and hidden drivers underpinning biological processes in health and disease. Ultimately, we aim to translate the hidden driver discoveries into therapeutic targets, biomarkers, and combination therapies for cancer and other disorders.

Our research summary
Our lab leverages expertise in computational and experimental biology, utilizes multi-omics technologies including single-cell and spatial omics, and collaborates with biologists, immunologists, and clinicians to explore the intracellular and intercellular networks within and between cells in complex systems and diseases (e.g., tumor and tumor microenvironment) and develop therapeutic strategies targeting the underlying hidden drivers to improve patient outcomes.
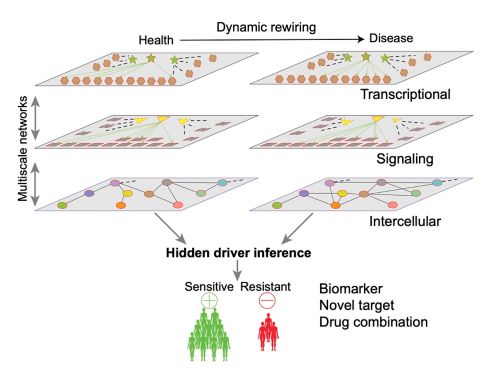
Hidden Driver Inference
Signaling, epigenetic, and metabolic factors are crucial drivers of network rewiring, and likely therapeutic targets in cancer, but are difficult to identify as they are commonly altered by posttranslational modifications or other mechanisms, thus referred as “Hidden drivers”. We have developed the NetBID algorithm to infer hidden drivers by integrating transcriptomics with other omics data, which identified Hippo kinase MST1 as a hidden driver of CD8a+ dendritic cells for antitumor immunity. NetBID has been widely used in basic biology and oncology. For example, by using NetBID, we discovered that LCK is the hidden driver of unexpected dasatinib sensitivity in >40% of pediatric T-ALL and intra-tumoral heterogeneity of T-ALL is characterized by T-cell development stages conferring distinct therapeutic vulnerabilities to dasatinib and venetoclax driven by a dynamic LCK–BCL2 switch.
Single-cell and Spatial Systems Biology
To decipher wiring and rewiring within and between individual cells and pinpoint underlying hidden drivers, we utilize single-cell and spatial omics technologies. We are developing a mutual information–based framework, scMINER, for clustering analysis, cell type–specific inference of intracellular networks, hidden drivers and network rewiring, and inference of intercellular communication networks. With spatial omics data, we are developing machine learning algorithms to refine cell-cell communication networks inferred from single-cell data and project hidden drivers into space. We have direct access to various single-cell and spatial omics technologies, including 10x Genomics (scRNA-seq, scATAC-seq, scMultiome, Visium), NanoString (GeoMx, CosMx), and CODEX.
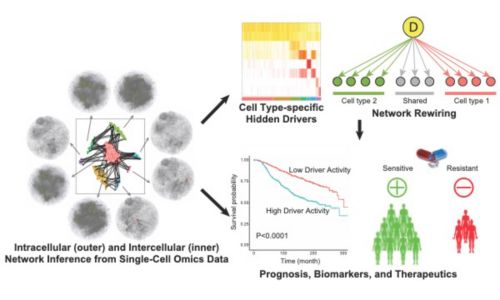
Systems Immunology and Immuno-Oncology
Together with Drs. Hongbo Chi, Terrance Geiger, and Stephen Gottschalk, we are co-leading the St. Jude iTARGETS Blue-sky Initiative (Immuno-Oncology Target Identification via Systems Immunology). The goal is to identify and translate innovative targets in immuno-oncology, ultimately enabling and advancing curative therapies for pediatric cancers and other catastrophic diseases. In particular, we are reconstructing the tumor and tumor microenvironment interaction maps by integrating bulk, single-cell, and spatial omics technologies for novel immunotherapy target identification in pediatric cancers. We are also working to identify and target hidden drivers to overcome CAR-T resistance, improve CAR-T persistence, and develop potential combination strategies with immune checkpoint blockades in pediatric cancers, especially pediatric solid and brain tumors.

Translational Systems Biology
The hidden drivers and activity from gene expression data could become the next generation targets and biomarkers that go beyond genomics for precision oncology. We work closely with cancer biologists, chemical biologists, and oncology clinicians to translate our systems biology discoveries of hidden drivers and networks into development of potential therapeutics and identification of relevant biomarkers. Our discoveries have been translated into metastatic breast cancer clinical trials (e.g., NCT02632071). The LCK-BCL2 discovery in T-ALL is being translated into multiple clinical trials across the world, including St. Jude, MD Anderson, EU, and Asia. We are developing the next generation activity-based biomarkers for targeted therapies. We are also developing combination therapies that target hidden drivers in different tumor clones, named clonal therapy (U01CA264610) or target tumor-TME interactions to minimize drug resistance, relapse, and metastasis for cancer treatment.

Selected Publications
Contact us
Jiyang Yu, PhD
Associate Member, Computational Biology
Department of Computational Biology
MS 1135, Room IA6051
St. Jude Children's Research Hospital

Memphis, TN, 38105-3678 USA GET DIRECTIONS